One of the main tasks of computational genetics is the identification of disease-associated genes. Case-control studies represent a crucial approach to addressing this challenge. To this end, a cohort of patients with the disease under study is established, in addition to a control group. Thereafter, by comparing the mutations observed in the DNA of each group and employing special statistical techniques, an association study is conducted. The outcome is a list of individual mutations or genes that are significantly associated with disease risks.
Case-control studies have been successfully applied to a very wide range of conditions, leading to the discovery of specific genes involved in disease risk and the identification of drug targets that have subsequently become medicines. It is noteworthy that participants in good health and without known congenital diseases are usually recruited as a control group, making them suitable for research on any disease. This approach would result in significant time savings in data collection and the avoidance of the need to generate genomic data multiple times.
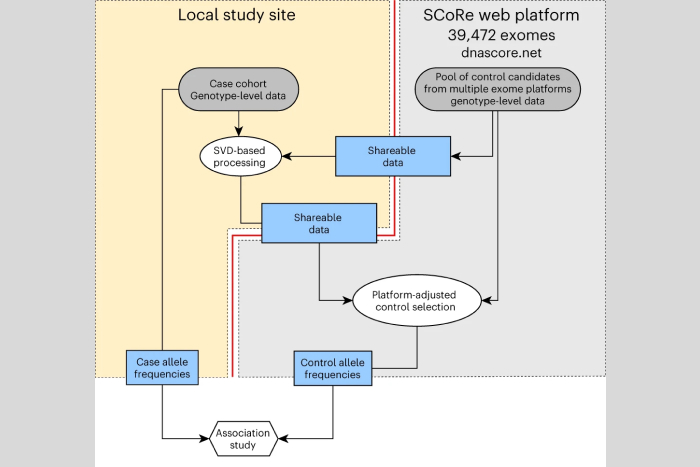
The concept of utilizing a shared «pool of controls» is not novel and has been discussed in genetic research for over a decade. However, the practical implementation of such a solution is associated with significant technical and ethical challenges. Genetic data is inherently personal, which means that the sharing of such data between research groups must comply with complex regulations on the protection of personal data, which requires considerable time and effort. If only generalized information about the control group – allele frequencies for DNA variants – is used, there is a systematic error in comparison with the study patient group, caused by differences in genetic ethnicity, technical features of sequencing and genomic data processing.
A joint study conducted by scientists from the WCRC for Personalized Medicine (St. Petersburg, Russia), Nationwide Children's Hospital (Columbus, USA), Broad Institute (Cambridge, USA), and Washington University (St. Louis, USA) resulted in the development of an algorithm for the selection of a control group matching the genetic features of the patient cohort under study without the use of personal data. This method does not involve the transfer of personal data, therefore, it is completely safe for study participants and does not require additional approvals. Furthermore, numerous technical aspects of association studies without the transfer of individual data were resolved during the course of the work. A control exome sequencing sample pool comprising 40,000 participants has been created, and a platform, SCoRe, has been developed for the purpose of selecting control samples for any patient group under study and for obtaining generalized genomic data of controls for subsequent use in association analysis.
The results of the study were published in the leading journal Nature Genetics https://www.nature.com/articles/s41588-023-01637-y in February 2024.
